Project Proposal: Fractal Dimension Measurement for Skin Cancer Imaging
Background
Dermoscopic skin imaging is a powerful tool for the early diagnosis and prognosis of skin cancer (melanoma). This imaging modality produces high-resolution images of skin structures such as naevi (moles). The image below shows such a structure.
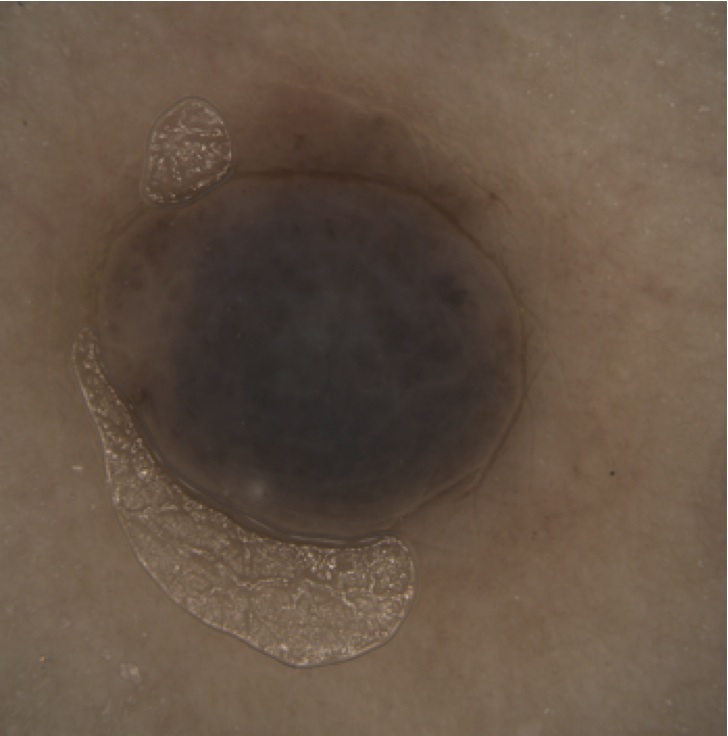
The malignity of such structures is typically measured using various parameters, commonly known as the ABCDE criteria (asymmetry, irregular borders, colors, diameter, and enlargement). Of these, border irregularity is particularly hard to measure.
Goal
In this project, we investigate the usage of image segmentation and shape quantification techniques for automatically measuring the border irregularity of skin structures. Border irregularity has earlier been quantified using a so-called fractal dimension. In this project, we aim to compute the border irregularity of naevi using a new technique: medial axes, or skeletons.
Workplan
The work is split into several steps, as follows
1. Image segmentation
To compute skeletons, we need first to extract shapes from the raw color dermoscopic images. For this, images need to be segmented. Appropriate segmentation criteria need to be defined, e.g. based on luminance and hue variation. The image below shows a simple luminance-based segmentation of the naevus in the top image. The border is shown in red. The grayscale background shows the so-called distance transform of the border.
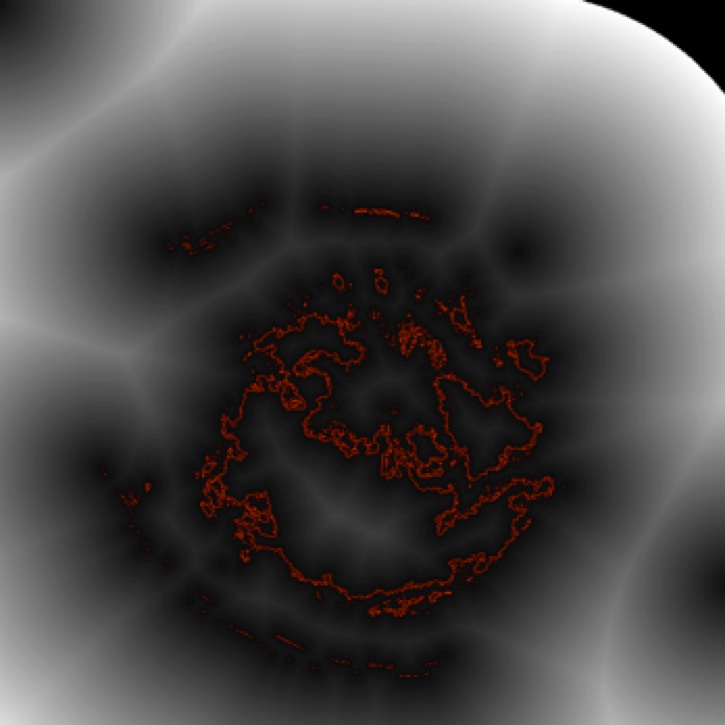
2. Skeletonization
For the segmented shapes, skeletons are computed. For this, one can use our skeletonization software available here. Appropriate clean-up of the extracted skeletons is needed e.g. to remove small-scale noise fragments. The image below shows, in white, the skeleton of the segmented red contour in the image above.
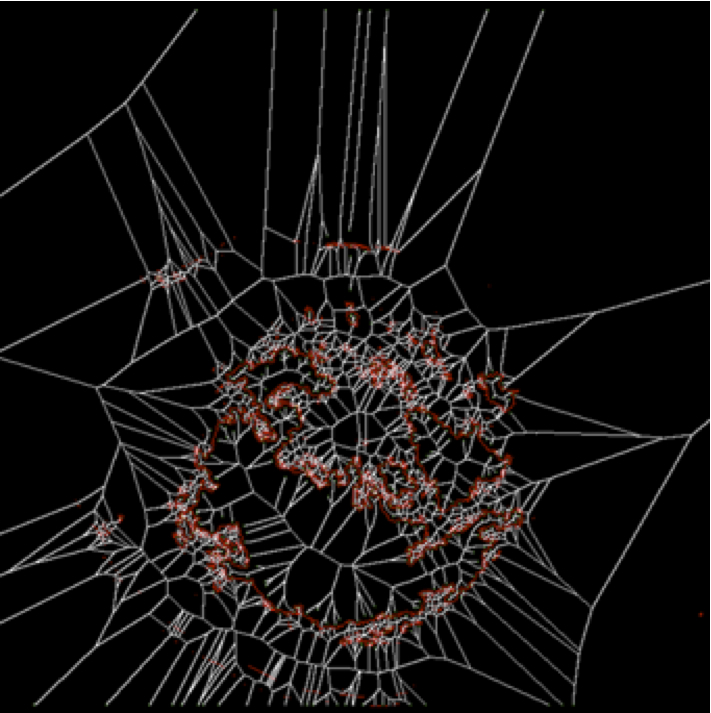
3. Border irregularity computation
Skeletons encode the irregularity of a shape's borders: The more jagged the border is, the more branch endpoints the skeleton will have. In this step, the number of endpoints of a given skeleton is computed. Appropriate clean-up is needed e.g. discarding the endpoints of very short, non-salient, branches.
4. Fuzzy shape detection
The above pipeline will not work well if the image segmentation is sensitive, as the same image may yield a multitude of shapes for different segmentation parameters. To regularize this, we propose to execute the pipeline steps 1..3 for a family of different segmentation parameters corresponding e.g. to different luminance or hue values. Helpful techniques in this area are max-trees. Given a family of shape contours, we can compute the fuzzy border irregularity as a histogram of the border irregularities of all contours in the family.
5. Comparison (optional)
To validate results, the entire pipeline is run on a set of predefined dermoscopic images. The resulting border irregularity values are next used to classify images into higly irregular and smooth-border. The classification results are compared with existing methods for measuring border irregularity, and with human insights from dermatologists. If the new method produces similar classifications and/or classifications in line with the human insights, we have succeeded in producing a good automatic image analyzer. This step is only to be done if sufficient time remains in the project.
Deliverables
- operational software implementing the imaging pipeline described above
- report (20..30 pages) describing the implemented algorithms and results
- results (tables with measurements on a set of predefined images, and the results of all processing steps on these images)
Provisions
The following material is already available
- robust, fast, easy-to-use skeletonization source code (C++)
- set of acquired dermoscopic images
- support from medical dermatologists in terms of evaluating results
Requirements
The candidate(s) for this project should ideally have
- good software development experience (C, C++, C# or Java)
- good analytical skills
- some practical and theoretical experience with image processing
Evaluation
The project will be evaluated as follows
- analysis of problem and solution space (completeness, clarity, justification; 30%)
- quality of produced imaging results (robustness to noise, classification matching human insight and/or existing classification metrics; 30%)
- quality of final report (completeness, readability, clarity; 20%)
- quality of implementation (ease of use, code documentation; 20%)
Participants
The project can be done by a single student or two students. If done by two students, then
- step 5 becomes mandatory
- work can be divided as follows
- student 1: steps 1,2,3
- student 2: steps 4,5
Literature
Several starting papers about evaluating skin tumors are available here.
Contact person
This project is supervised by prof. Alexandru Telea.
The second supervisor is dr. Michael Wilkinson.