Mathematical Computer Modelling of Host and Microflora Interactions
Contents of this File
- General Remarks
- Model Intestinal Microflora in Computer Simulation
- Current work
- Future improvements
- References
1. General Remarks
Computer simulation and mathematical modelling are not frequently used in the study of the
intestinal microflora, even though the first models date from the early 80s
(Freter et al. (1983). Given the complexity of the intestinal
microflora, developing theories which are consistent with in vitro data and are
at least self-consistent is probably impossible without a more rigorous mathematical framework
backed up by computer simulations. The former provide sthe basis for the latter, and the
latter provide rigorous tests for "thought experiments". However, given the same complexity of the microflora, providing such mathematical and computer models is daunting, and a careful, step-by-step approach is needed.
One use of such models is checking whether
combined in vitro data obtained from, e.g., chemostat experiments can
indeed mimic the in vivo behavior of the complete system.
Furthermore, as has been done with the
Cybermouse in silico
murine immune system, models can be used both to design in vivo
experiments rationally and to assist in the interpretation of results. Before
any experimentation begins, parameters which give the best discrimination
between competing theories may be determined. This may lead to a reduction of
the number of laboratory animals, healthy volunteers and patients needed.
2. Model Intestinal Microflora in Computer Simulation
(MIMICS)
The MIMICS project is the first attempt to simulate the intestinal microflora using
supercomputers (Wilkinson, 1997;
Kamerman & Wilkinson, 2002; Wilkinson, 2002).
A computer program has been developed to simulate the intestinal microflora. Initially, the
ISGNAS funded a study to explore the uses and feasibility of the development of computer
models in the study of factors influencing the microbial ecology, and in particular the
colonization resistance of
the digestive tract.
The results(Wilkinson, 1997) of
the ISGNAS-pilot study show that modelling of interactions within the gut
microbial ecosystem is feasible.
NOTE: What is unlikely to be accomplished with such a model is prediction of developments in the microflora of individual patients. The number of unknown and unmeasurable parameters is probably too high to ever achieve such accuracy in modeling.
3. Current work
Adding epithelial binding sites is currently being done (Kamerman & Wilkinson, 2002), much along the lines of the model of Freter et al.
(1983). Furthermore, bacteriocin-based interactions are being modeled,
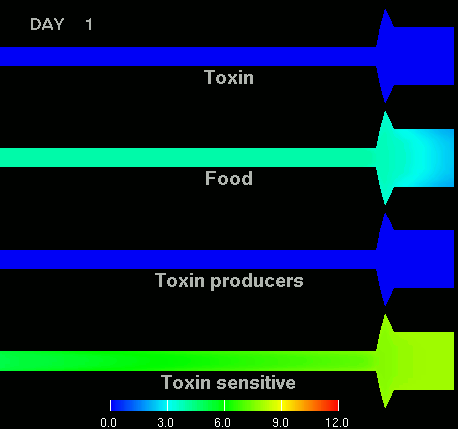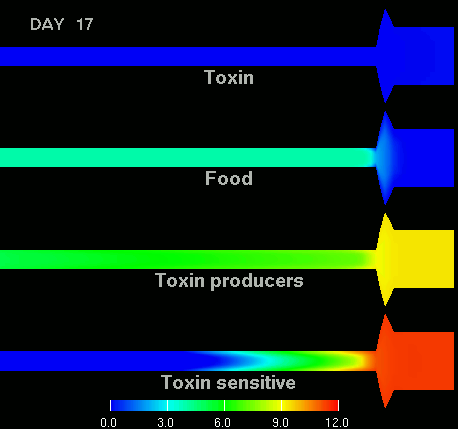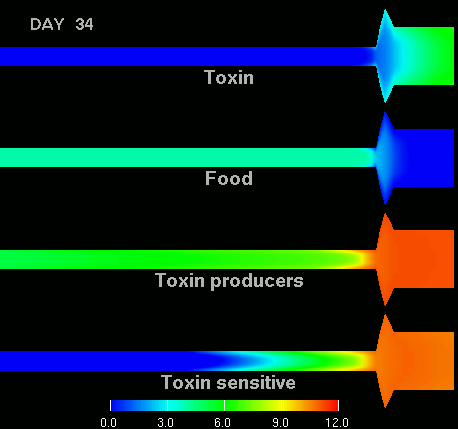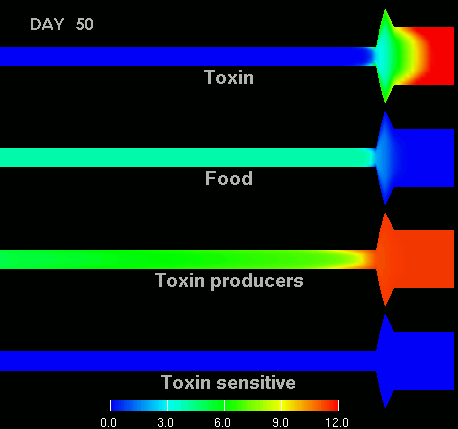
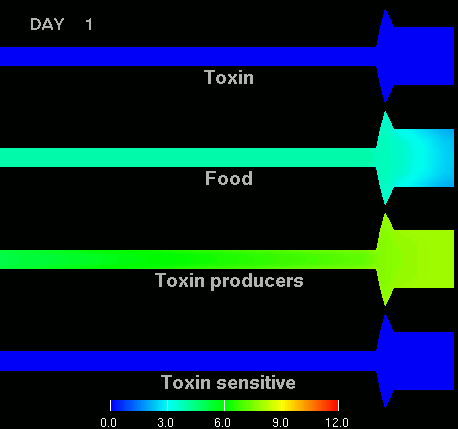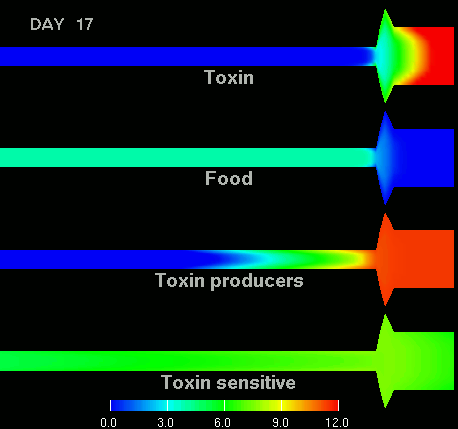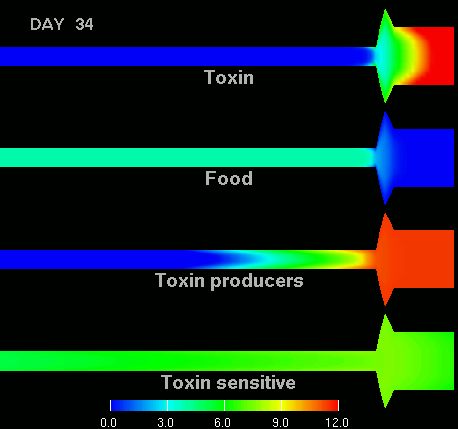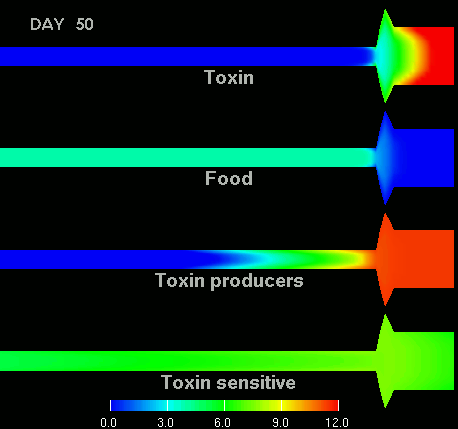
as shown in the figure below.
(Left) After 7 days of colonization of the intestine by a bacteriocin
(=antibacterial toxin) producing strain of bacteria it is impossible to
replace them by toxin sensitive bacteria, even if they are ingested every day
for fifty days. (Right) Note difference with when a bacteriocine producing bacterium
arives after colonization by toxin sensitive species. The unit of the scale is 10log of the number of bacteria/g. The length of the intestine is 6 m, the diameter varies from 3 cm in the small intestine through 10 cm in the cecum, back to 6 in the colon. Radial scale has been exagerated for display purposes.
See also this article on this simulation.
4. Future improvements
The model as it stands in far from complete. A step-by-step plan to improve the model is proposed:
- Add more different species of bacteria
- Add a wider variety of food substrates and toxins
- Allow bacteria to produce metabolites
- Combine to form food webs
- Add chemotaxis
- Within this more complex model, study CR.
- Add an immune system
The current model already supports bacterial motility (without chemotaxis), by increasing the diffusion rates of certain species at the expense of growth rate (higher basal metabolism). For the first four steps, a more or less generic (meta)model of a bacterial metabolism must be made. Once this has been formulated it can de used in two very different ways: (i) tactically, to closely mimic existing bacteria by careful design, and to see whether this models the actual system, and (ii) strategically, by assigning random metabolisms (within certain reasonable limits) to species, entering various combinations of substrates, and to see what type of food web emerges, and which types of survival strategies are feasible within this artificial environment. Evolution could also be mimicked by introducing random changes into the metabolic properties of bacteria. It is obvious that there are many possible in silico experiments within the current framework. An invitation is issued to researchers within the field of study
of the ISGNAS to design and if possible conduct such experiments in collaboration with the ISGNAS effort.
The inclusion of an immune system should involve the use of one or more of
the in silico model immune systems developed elsewhere (e.g. the
Theoretical
Immunology Group at
SFI). Links with competing
models could provide a valuable test of the applicability of each model.
References
- Freter R, Brickner H, Fekete J, Vickerman MM, Carey KV (1983) Survival and implantation of Escherichia coli in the intestinal tract. Infect. Immun. 39:686-703.
- D.J. Kamerman and M.H.F. Wilkinson (2002) In silico modelling of the
human intestinal microflora. Lecture Notes in Computer Science Vol. 2329 Proc. ICCS 2002, (P.M.A. Sloot et al eds), Amsterdam, The Netherlands, pp. 117-126. PDF (201 kB) preprint version available.
- Wilkinson MHF (1997)
Nonlinear dynamics, chaos-theory, and the "sciences of complexity":
their relevance to the study of the interaction between host and microflora
In: "Old Herborn University Seminar Monograph Vol. 10: New Antimicrobial
Strategies"
(P.J. Heidt, V. Rusch, and D. van der Waaij, eds.), pp. 111-130, Herborn Litterae,
Herborn-Dill, Germany.
PDF (842 kB) version available.
- M.H.F. Wilkinson (2002) Model intestinal microflora in computer
simulation: a simulation and modelling package for host-microflora interactions.
IEEE Trans. Biomed. Eng. 49:1077-1085.
PDF (417 kB) preprint version available.
Send comments and suggestions to
M.H.F. Wilkinson