Project Proposal: Root Plant Skeletonization
Background
Biology applications rely increasingly more on data acquisition techniques such as 3D scanning. Such scans, for example, can contain 3D volumetric representations of a plant, e.g. the root structure. Much like in medical applications (CT,MRI) scanned datasets can be processed to quantify various aspects of the scanned data. For plants, these can be the geometry and topology of the root structure, e.g. number of branches of a given size, ramification factor, branch twists, maximal/average branch thickness, etc.
Goal
In this project, we develop new automatic methods for the extraction of relevant geometric and topological metrics from plant root scans. As input, we have a 3D grayscale volume representing e.g. the type of 'tissue' present at a given location (voxel) in the plant. The volume is obtained from a synthetic 3D reconstruction of the plant root using a sequence of 2D images. Ultimately, we want to compute several metrics describing the plant root structure, such as
- distribution of lengths of the root branches
- ramification factor of the branches (how/where/how often do side-branches emerge)
- variation of thickness of branches along their main path
Skeletonization
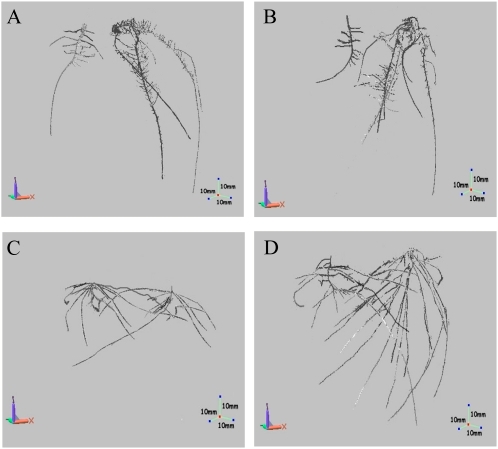
A typical first step when reasoning about the structure of a branch-rich structure embedded in a 3D volume is to compute the so-called skeleton or medial axis of the scanned shape. The image below shows example skeletons of several plant root structures.
For this, we propose using a fast and robust 3D skeletonization method available here.
However, computing a raw skeleton of a shape is not enough to quantify the shape. Several operations are required additionally to that
- eliminate small-scale noise fragments (e.g. small connected components and/or small branches)
- clean up the emerging skeleton in order to compute a one-voxel-thin, connected, skeleton
- identify the tips (endpoints) of the skeleton
- compute paths and path-lengths in the emerging skeleton graph
- visualize the computed structures (skeletons, paths, metrics) in an easy-to-understand manner
Workplan
The workplan comprises the following steps
1. Related work study
In this step, related work on the computation of skeletons from voxel datasets is studied. The outcome should be an already-implemented, robust, fast, scalable method to compute such skeletons from relatively large 3D volumes (500^3 voxels). In particular, the method and code available here should be tried.
2. Clean skeletons
In this step, skeletons are computed from the input images. The skeletons are, if needed, postprocessed to eliminate noise, disconnections (where possible), and make them one-voxel-thin.
3. Skeleton graph
In this step, the skeleton is processed to find its endpoints, paths between endpoints, and path-related metrics. This may require the conversion of the voxel skeleton into a graph and various (adapted) graph traversal algorithms.
4. Visualization
In this step, a (simple) visualization application is built to allow users to interactively visualize the computed results and/or perform various computational tasks interactively.
Deliverables
- operational software implementing the skeletonization tool described above
- report (20..30 pages) describing the implemented algorithms and results
- results (tables with measurements on a set of 3D scans, and the results of all processing steps on these images)
Provisions
The following material is already available
- robust, fast, easy-to-use, general-purpose skeletonization source code (C++)
- 3D scanned datasets of plant roots in a simple-to-read format
Requirements
The candidate(s) for this project should ideally have
- (very) good software development skills (preferably C/C++)
- good analytical skills
- image processing skills are an extra advantage
Evaluation
The project will be evaluated as follows
- analysis of problem and solution space (completeness, clarity, justification; 30%)
- quality of produced imaging results (robustness to noise, measurements matching the human insights, speed; 30%)
- quality of final report (completeness, readability, clarity; 20%)
- quality of implementation (ease of use, code documentation; 20%)
Participants
The project can be done by a single student.
Literature
Many papers on 2D and 3D skeletonization are available navigating the skeleton-related project pages available here.
Contact person
This project is supervised by prof. Alexandru Telea.
The project is in cooperation with prof. Alexandre Falcão (Univ. of Campinas, Brazil), and with prof. Leon V Kochian (Robert W. Holley Center, Cornell University, USA).