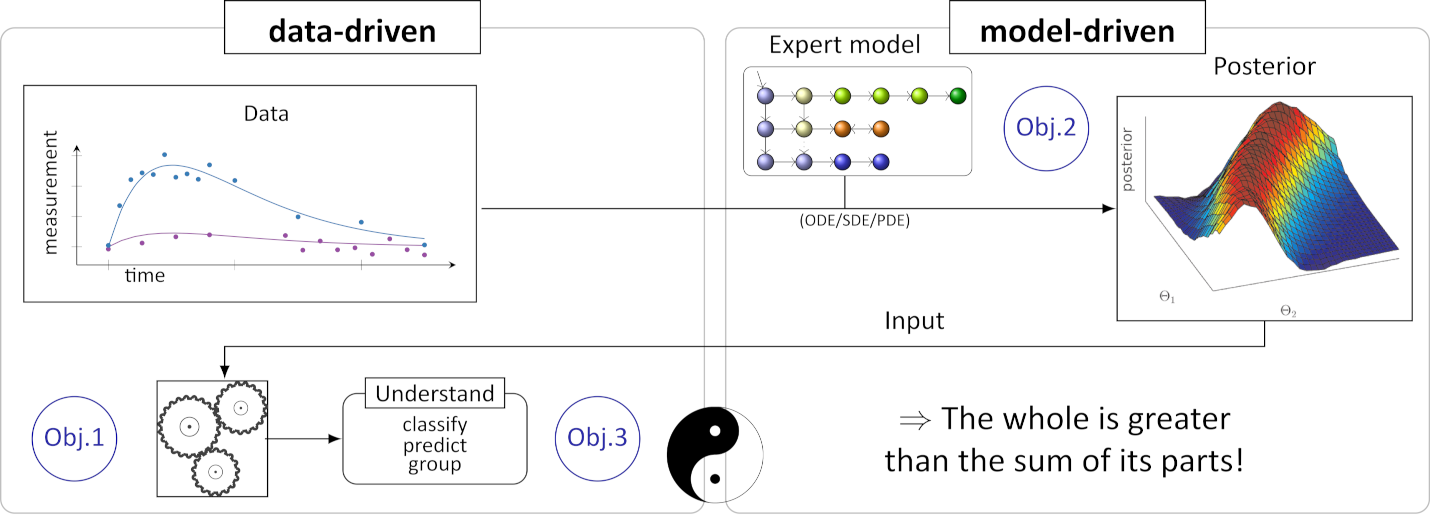
Nowadays, most successful machine learning (ML) techniques for the analysis of complex interdisciplinary data use significant amounts of measurements as input to a statistical system. The domain expert knowledge is o en only used in data preprocessing. The subsequently trained technique appears as a “black box”, which is difficult to interpret and rarely allows insight into the underlying natural process. Especially in cri cal domains such as medicine and engineering, the analysis of dynamic data in the form of sequences and me series is o en difficult. Due to natural or cost limitations and ethical considerations data is o en irregularly and sparsely sampled and the underlying dynamic system is complex. Therefore, domain experts currently enter a me-consuming and laborious cycle of mechanistic model construction and simula on, o en without direct use of the experimental data or the task at hand.
We published a hybrid approach were proposed combining the predictive power of ML and the explanatory power of pharmacokinetic models for model-based clustering, automatically determining groups of responses to medication in a clinical data set. In this VIDI-project, we aim to formulate a generalized framework for learning in the space of dynamic models that represent the complex underlying natural processes, with potentially very few measurements. This will be achieved by incorporating expert knowledge in the form of mechanistic models and their structural properties and task-driven model reduction.